Engineering Cell Fate (aka reprogramming)
During development, cells change their gene expression program and form specific cell types that become increasingly resistant to cell fate changes. Thus, differentiated cell states are remarkably stable in vivo. However, they can be reprogrammed to the pluripotent state, or into other lineages, by overexpression of only a few transcription factors or through combinations of chemicals. Our long-term goal is to understand the fundamental mechanisms that enable these cell fate transitions.
We are well known for our pioneering work on somatic cell reprogramming to induced pluripotent stem cells (iPSCs). Our work includes the derivation and characterization of fully reprogrammed mouse iPSCs (Maherali et al, Cell Stem Cell, 2007), the establishment of human iPSCs (Lowry et al, PNAS, 2008), the characterization of the epigenetic state of mouse and human iPSCs (Maherali et al, Cell Stem Cell, 2007, Tchieu et al, Cell Stem Cell, 2010, Chin et al, Cell Stem Cell, 2009, Chin et al, Cell Stem Cell, 2010, Chronis et al, Cell, 2017), the identification of barriers of the reprogramming process (Ho et al, Cell Reports, 2013, Sridharan et al, Nature Cell Biology, 2013) and of reprogramming steps (Pasque et al, Cell, 2014), the elucidation of reprogramming factor action (Sridharan et al, Cell, 2009, Chronis et al, Cell, 2017), and the use of the inactive X chromosome to reveal insights into the chromatin reorganization during the reprogramming of human and mouse somatic cells (Maherali et al, Cell Stem Cell, 2007, Pasque et al, Cell, 2014, Tchieu et al, Cell Stem Cell, 2010). The ability to generate all cell types of the body has thrust iPSCs into the spotlight as a panacea for regenerative medicine. With the generation of human iPSCs it became feasible to generate pluripotent cells from any person by reprogramming, which has ushered in an era of personalized medicine and “disease-in-a-dish” modeling.
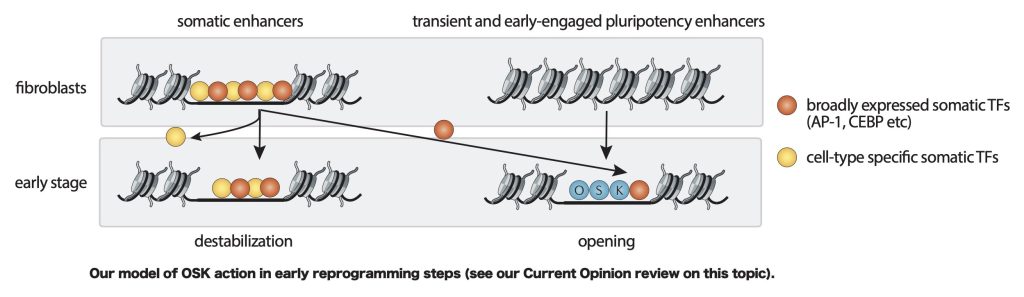
A key focus of our lab has been on understanding how the reprogramming factors Oct4, Sox2, Klf4 and cMyc (OSKM) can induce pluripotency in somatic cells. We showed that OSK mediate the cell fate change of reprogramming by activating pluripotency enhancers through collaborative binding. A critical observation we made was that early in reprogramming, somatic enhancers are perturbed genome-wide and display decreased levels of active enhancer marks, decreased chromatin accessibility, and decreased binding of somatic transcription factors (TFs) without a dramatic change in somatic TF expression levels. We found that this global destabilization of somatic enhancers is not driven through their direct interaction with OSK and instead with the redistribution of somatic TFs to newly opened sites bound by OSK. These new sites carry canonical motifs for OSK and somatic TFs. The most parsimonious model explaining somatic enhancer inactivation therefore is that OSK redirect somatic TF binding by recruiting them to their target sites in newly opening enhancers and simultaneously removing them from somatic enhancers, leading to widespread somatic gene silencing. Since OSK thereby induce the redistribution of widely expressed and critical somatic TFs such as AP1, our model provides an explanation for how OSKM can induce iPSCs in many diverse somatic cell types. Overall, our findings set the stage for us to fully understand how to manipulate cell identity and rationally alter cell states (Deng, Jacobsen, Collier et al Current Opinion 2021).
Current directions in the lab include:
- Can we use CRISPRi to perturb enhancers and genes to build a comprehensive catalog of critical gene regulatory programs for reprogramming to iPSCs and pluripotent cell conversions?
- Connecting Variants to Function: Can we advance human genetics by building catalogues of reprogramming epigenomes for a large cohort of human cells to understand how genetic differences alter the epigenome and thereby human health?
- Can we understand the mechanisms underlying rejuvenation by OSKM?
- Can we reconstruct cell histories during reprogramming?