Inventing New Technologies
At the heart of our research group, we apply and develop sequencing technologies to better understand gene regulation and cell function across both health and disease. In previous work, we have u Chip-seq, RNA-seq, 4C-seq, 4C-seq, Hi-C, CnR/T, RAP-seq, and ATAC-seq and utilize many of these approaches to profile single cells to address the questions we are interested in. We also collaborate with labs at UCLA and beyond to apply our genomics approaches to normal and disease tissues. Pushing the boundaries of single cell genomics, we are now leveraging spatial genomics techniques to map the transcriptomes of individual cells in tissues. In addition, create innovative computational approaches to analyze our genomics data. We are also using metabolomics and proteomics as well as sophisticated imaging approaches.
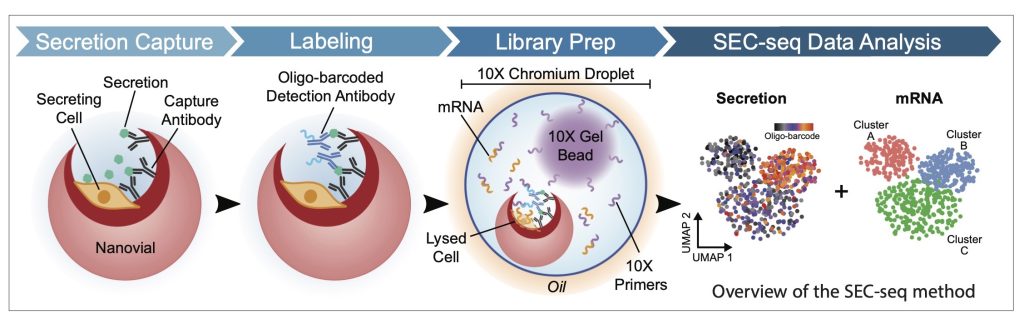
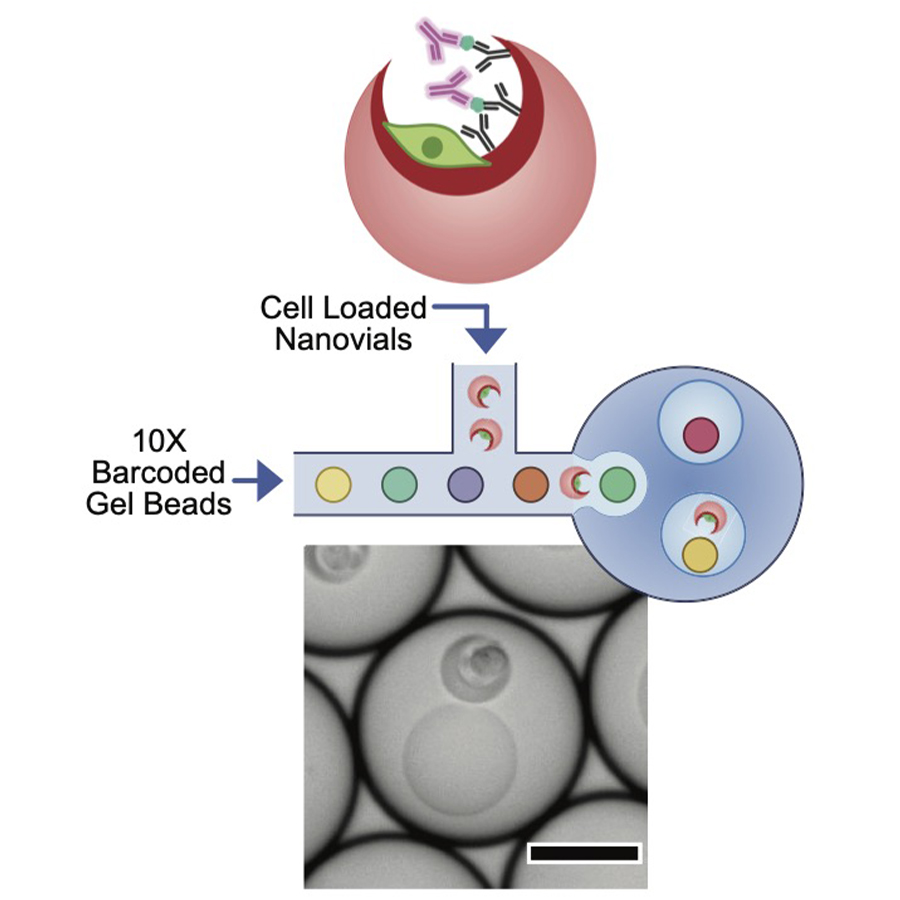
As an example, collaborating with the DiCarlo lab at UCLA, we developed a new method that enables the simultaneous measurement of the transcriptome and secreted proteins (SEC-seq). Secretion Encoded Single-Cell sequencing (SEC-seq) (Udani, Langerman et al, Nature Nano, 2024) leverages microscale hydrogel particles (nanovials) and single-cell (sc) RNA-seq in microfludic droplet emulsions, to retain and analyze both the transcriptome and secretion information from individual cells. Briefly, to achieve SEC-seq, cells are loaded into gelatin-coated nanovials conjugated with capture antibodies for a secreted protein of interest, allowing the cells to adhere to the nanovial surface and secreted protein to bind to the capture antibodies. After a short incubation time, nanovials are incubated with oligonucleotide (oligo)-barcoded detection antibodies against the secreted product. Single-cell loaded nanovials, enriched by FACS, are then partitioned for downstream scRNA-seq, followed by library preparation for mRNA and oligo-barcode detection (for measuring detection antibody molecules), sequencing, and data analysis.
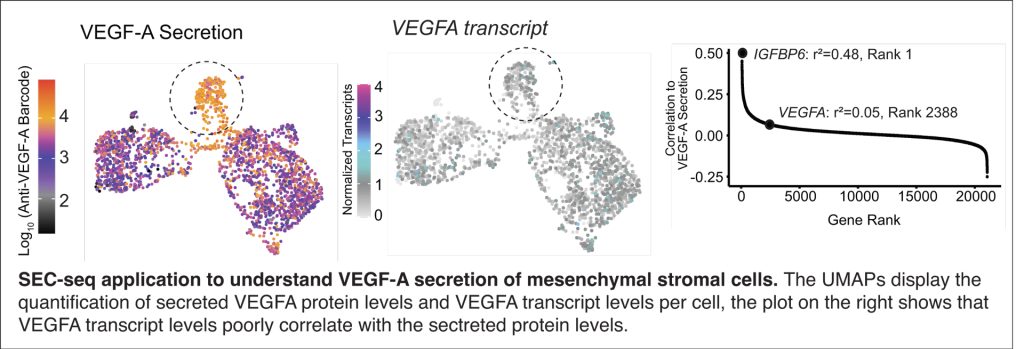
Over 3000 proteins are predicted to be secreted from human cells, and secreted proteins such as immunoglobulins, cytokines, chemokines, extracellular matrix proteins, proteases, morphogens, and growth factors span a diversity of critical functions. For example, mesenchymal stromal cells (MSCs) have been widely evaluated as therapeutics because they secrete bioactive factors including growth and neurotrophic factors, cytokines, and extracellular vesicles, which promote immunomodulation and regeneration. Yet, successful translation of MSCs and other cell therapies has been hindered by clinical outcome inconsistency attributed to functional differences that stem from the cell source and cellular heterogeneity. The exploitation of the full potential of cell therapies therefore necessitates an understanding of the sub-populations that secrete the proteins of interest; ideally detecting protein secretion at the single-cell level. SEC-seq allows us to define distinct secretory cell states through their gene expression state, enabling a thorough characterization of the secreting sub-populations and the identification of new regulatory mechanisms and required components in secretory pathways.
SEC-seq can easily be adopted by anyone who has access to current single-cell sequencing instruments, like the 10X Chromium and FACS, without the need for specialized microfluidics equipment and skills. The general workflow is modular, enabling researchers to investigate any secretion in which a pair of affinity reagents (i.e. an antibody pair for a sandwich immunoassay) is available. Overall, SEC-seq is opening the door to better cell-based therapeutics, new bioengineering approaches, and advancing the understanding of cell function.
Current directions in the lab include:
- Can we enhance the Sec-seq technology to simultaneously measure more secreted proteins?
- Can we apply Sec-seq to human developmental problems