Disease Mechanisms
We are interested in mapping disease processes and identifying pathways that contribute to diseases, with the aim of developing targeted therapies and treatments.
One interest of our lab are X-linked diseases. Characterizing female human iPSCs, we found that they exhibit nonrandom X-chromosome inactivation (Tchieu et al, Cell Stem Cell, 2010). Consequently, human iPSC lines derived from females with an X-linked disorder will express either the X carrying the mutant or the normal allele. Devastating X-linked genetic diseases include for example fragile X syndrome (mutation in FMR1), a-thalassemia (ATRX), Rett Syndrome (MECP2), Coffin-Lowry Syndrome (RSK2), DMD, Lesch-Nyhan syndrome (HPRT), and Wiskott Aldrich Syndrome (WASP). For studies of X-linked diseases with female human iPSCs, one therefore needs to carefully analyze which X chromosome is expressed. We derived Rett Syndrome iPSCs, which the Novitch lab at UCLA used create brain organoids. These organoids exhibited highly abnormal and epileptiform-like activity and are powerful in therapeutic drug discovery (Samarasinghe et al, Nature Neuroscience, 2021).
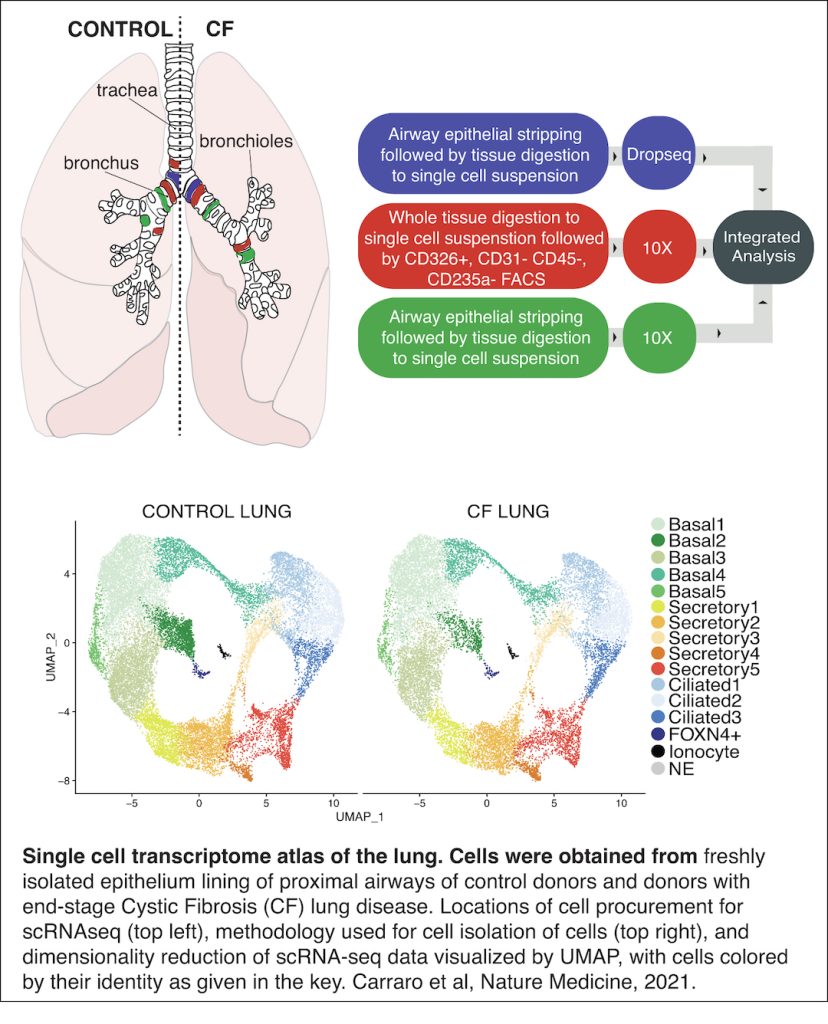
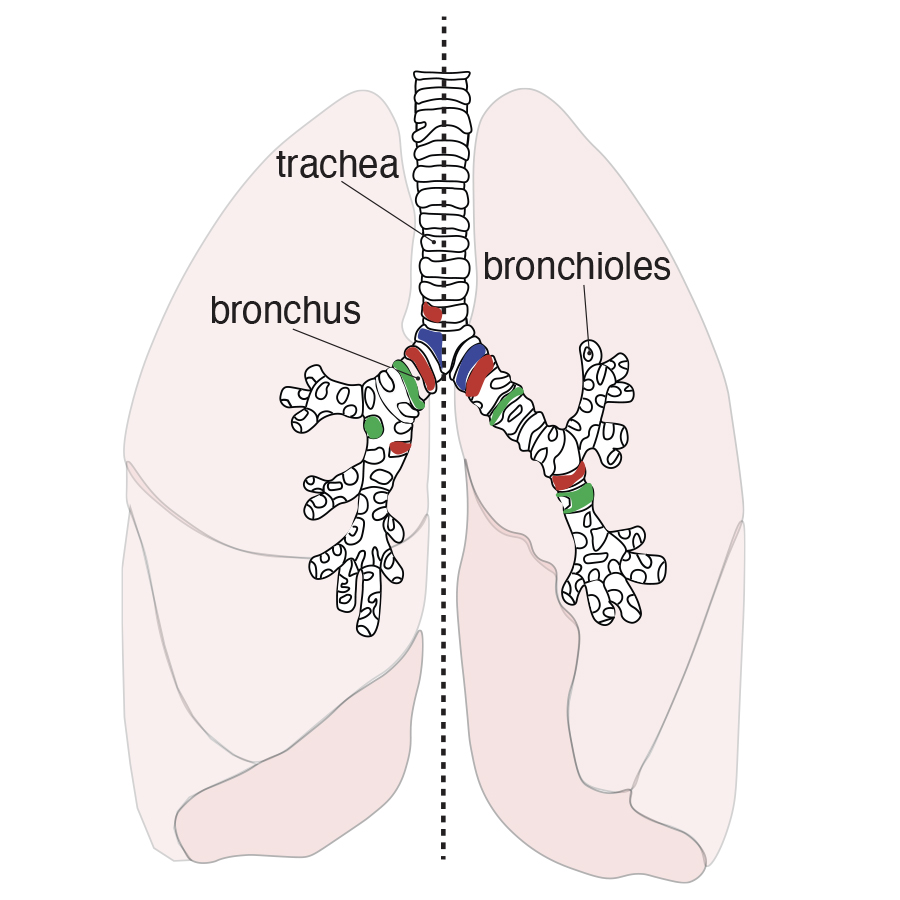
In addition to the work on the X chromosome, we extensively collaborate to apply our omics and computational approaches to understand human diseases. For example, we often work with the Gomperts lab at UCLA to study diseases of the lung such as Cystic Fibrosis. Cystic Fibrosis is progressive genetic disorder that affects more than 70,000 people worldwide, and results from mutations to the CFTR gene. Cells that contain the defective protein encoded by the gene produce unusually thick and sticky mucus that builds up in the lungs and other organs, clogging the airways, trapping germs and bacteria, causing life-threatening infections and irreversible lung damage. The Gomperts team and other collaborators collected lung tissue from patients; we developed a computational approach to compare the gene expressions patterns of the various cells, which allowed us to create a catalog of the cell types and subtypes present in healthy airways and those affected by cystic fibrosis, including previously unknown subtypes that illuminate how the disease alters the cellular landscape of the airways (Carraro et al, Nature Medicine, 2021). We are continuing the fruitful collaborations with the Gomperts lab and are participating in the NIH Lungmap consortium.
Current directions in the lab
- Can we improve tools to define cell states, understand how cell states are changing and infer trajectories in single cell omics data
- Analyze large scRNA-seq data sets from the lung
- Apply Sec-seq to the lung